library(tidyverse)── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorssick_data <- read_csv("sick-fish.csv")Rows: 1000 Columns: 12
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (1): species
dbl (10): tank_id, avg_daily_temp, num_fish, day_length, tank_volume, size_d...
lgl (1): below
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.glimpse(sick_data)Rows: 1,000
Columns: 12
$ tank_id <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16…
$ species <chr> "tilapia", "tilapia", "tilapia", "tilapia", "tilapia"…
$ avg_daily_temp <dbl> 22.95922, 23.98088, 23.97097, 24.26474, 24.29623, 23.…
$ num_fish <dbl> 95, 96, 101, 98, 93, 101, 98, 109, 97, 102, 99, 99, 9…
$ day_length <dbl> 9, 11, 11, 10, 10, 11, 12, 10, 10, 10, 9, 11, 11, 10,…
$ tank_volume <dbl> 399.6975, 399.8071, 398.8427, 399.8410, 399.7561, 398…
$ size_day_30 <dbl> 2784.895, 2781.003, 2785.807, 2785.253, 2786.946, 278…
$ ammonia <dbl> 0.10561057, 0.09073854, 0.10867733, 0.09421766, 0.093…
$ avg_daily_temp_F <dbl> 73.32660, 75.16558, 75.14774, 75.67654, 75.73322, 75.…
$ below <lgl> TRUE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE…
$ num_sick <dbl> 63, 18, 12, 0, 5, 11, 11, 0, 55, 23, 7, 65, 61, 62, 6…
$ oxygen <dbl> 9.480023, 9.288952, 9.467007, 9.322897, 9.327849, 9.4…sick_data <- sick_data %>%
mutate(perc_sick = num_sick/num_fish)
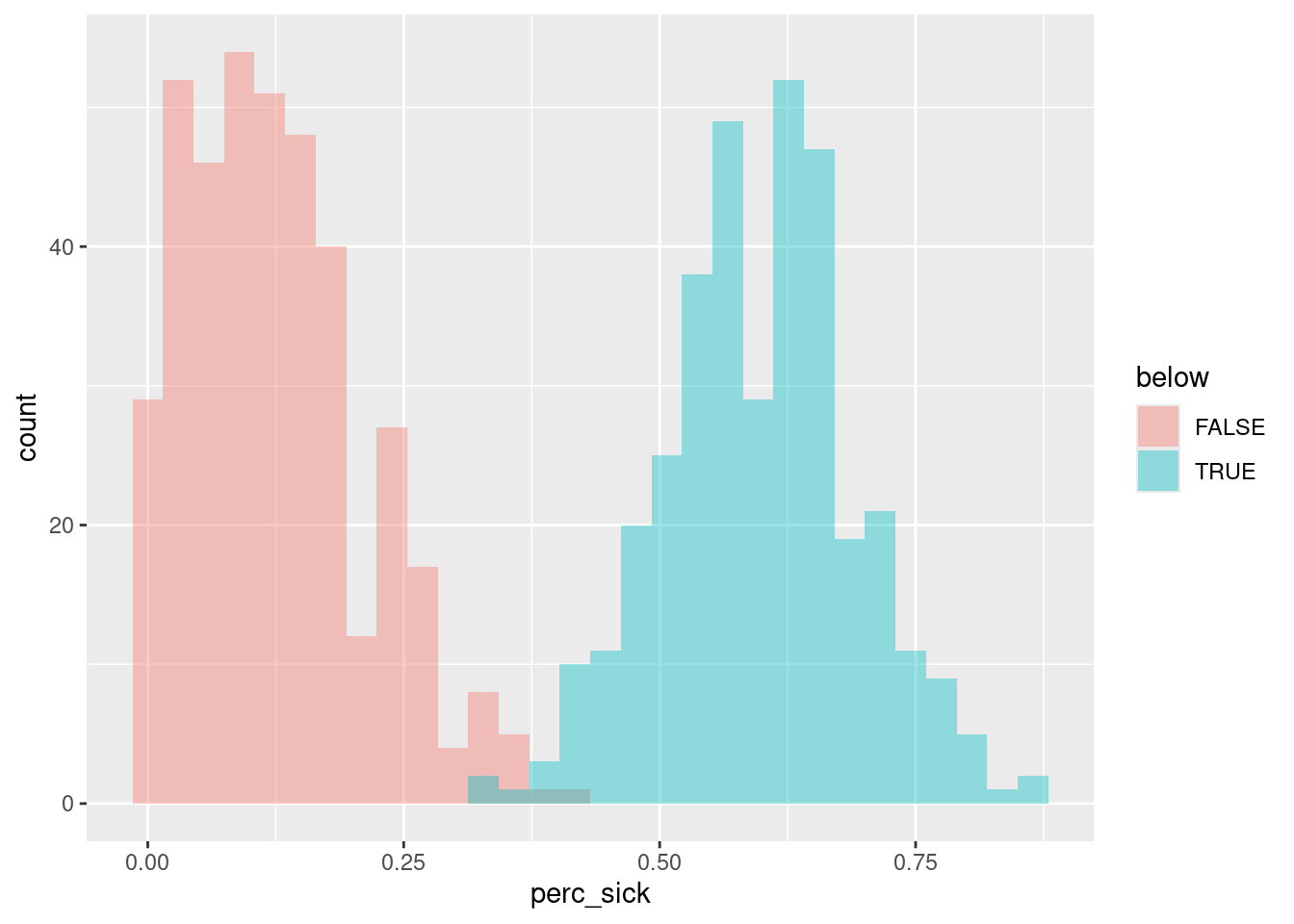
sick_data %>%
filter(species=="tilapia") %>%
group_by(below) %>%
summarize(mean = mean(perc_sick))# A tibble: 2 × 2
below mean
<lgl> <dbl>
1 FALSE 0.125
2 TRUE 0.597sick_data %>%
filter(species=="tilapia") %>%
ggplot(aes(x=perc_sick, fill=below)) +
geom_histogram(alpha=0.4, position = "identity")`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.